Main Article Content
Abstract
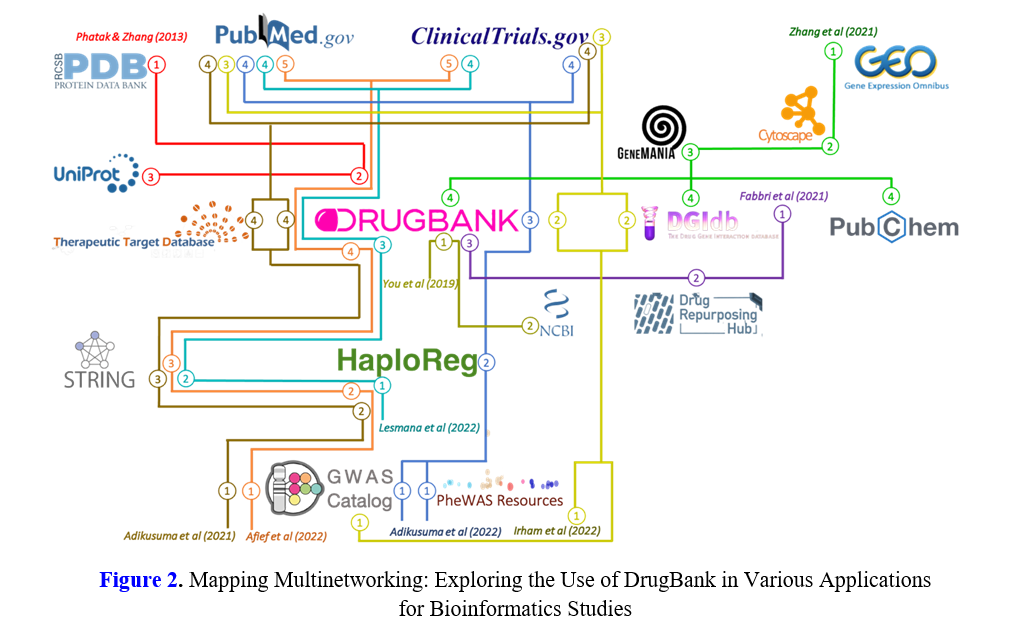
Bioinformatics plays a vital role in drug discovery and repurposing, yet challenges persist in data availability, biological complexity, and method standardization. The continued exploration and utilization of resources such as DrugBank will play a crucial role in advancing drug development and uncovering novel therapeutic opportunities. Our study presents a comprehensive analysis of the methodology utilized by bioinformaticians to integrate DrugBank with multiple databases, with the aim of facilitating the discovery of novel drugs. A literature review was conducted using Scopus and PubMed, focusing on articles from the last 10 years. Relevant articles meeting the inclusion criteria were collected between October and November 2022. The review identified 35 unique papers after removing duplicates. Screening led to 9 papers meeting inclusion criteria. The study reveals that DrugBank is an indispensable resource, aiding drug-gene interaction analysis and connecting gene data sources with potential drug candidates. It streamlines the multinetworking process and enables the identification and validation of new medications through clinical tools. These findings shed light on drug-gene interactions and drug repurposing, emphasizing the significance of leveraging multiple databases and network data. DrugBank's pivotal role in advancing drug discovery and personalized medicine underscores its importance in bioinformatics research.
Keywords
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
References
- Abiyana, S. R., Santoso, S. B., Pribadi, P., Hapsari, W. S., & Syarifuddin, A. (2024). From the Drugbank Application to the Novel Drugs: A Pharmacogenomic Summary. E3S Web of Conferences, 500, 04002. https://doi.org/10.1051/e3sconf/202450004002
- Adikusuma, W., Chou, W.-H., Lin, M.-R., Ting, J., Irham, L. M., Perwitasari, D. A., Chang, W.-P., & Chang, W.-C. (2022). Identification of Druggable Genes for Asthma by Integrated Genomic Network Analysis. Biomedicines, 10(1), 113. https://doi.org/10.3390/biomedicines10010113
- Adikusuma, W., Irham, L. M., Chou, W.-H., Wong, H. S.-C., Mugiyanto, E., Ting, J., Perwitasari, D. A., Chang, W.-P., & Chang, W.-C. (2021). Drug Repurposing for Atopic Dermatitis by Integration of Gene Networking and Genomic Information. Frontiers in Immunology, 12, 724277. https://doi.org/10.3389/fimmu.2021.724277
- Afief, A. R., Irham, L. M., Adikusuma, W., Perwitasari, D. A., Brahmadhi, A., & Cheung, R. (2022). Integration of genomic variants and bioinformatic-based approach to drive drug repurposing for multiple sclerosis. Biochemistry and Biophysics Reports, 32, 101337. https://doi.org/10.1016/j.bbrep.2022.101337
- Balamurugan, S., Krishnan, A., Goyal, D., Chandrasekaran, B., & Pandi, B. (Eds.). (2021). Front Matter. In Computation in Bioinformatics (1st ed.). Wiley. https://doi.org/10.1002/9781119654803.fmatter
- Cha, Y., Erez, T., Reynolds, I. J., Kumar, D., Ross, J., Koytiger, G., Kusko, R., Zeskind, B., Risso, S., Kagan, E., Papapetropoulos, S., Grossman, I., & Laifenfeld, D. (2018). Drug repurposing from the perspective of pharmaceutical companies: Drug repurposing in pharmaceutical companies. British Journal of Pharmacology, 175(2), 168–180. https://doi.org/10.1111/bph.13798
- Challa, A. P., Lavieri, R. R., Lewis, J. T., Zaleski, N. M., Shirey-Rice, J. K., Harris, P. A., Aronoff, D. M., & Pulley, J. M. (2019). Systematically Prioritizing Candidates in Genome-Based Drug Repurposing. ASSAY and Drug Development Technologies, 17(8), 352–363. https://doi.org/10.1089/adt.2019.950
- Cong, Y., Shintani, M., Imanari, F., Osada, N., & Endo, T. (2022). A New Approach to Drug Repurposing with Two-Stage Prediction, Machine Learning, and Unsupervised Clustering of Gene Expression. OMICS: A Journal of Integrative Biology, 26(6), 339–347. https://doi.org/10.1089/omi.2022.0026
- Cotto, K. C., Wagner, A. H., Feng, Y.-Y., Kiwala, S., Coffman, A. C., Spies, G., Wollam, A., Spies, N. C., Griffith, O. L., & Griffith, M. (2018). DGIdb 3.0: A redesign and expansion of the drug–gene interaction database. Nucleic Acids Research, 46(D1), D1068–D1073. https://doi.org/10.1093/nar/gkx1143
- Davies, M., Nowotka, M., Papadatos, G., Dedman, N., Gaulton, A., Atkinson, F., Bellis, L., & Overington, J. P. (2015). ChEMBL web services: Streamlining access to drug discovery data and utilities. Nucleic Acids Research, 43(W1), W612–W620. https://doi.org/10.1093/nar/gkv352
- Fabbri, C., Kasper, S., Zohar, J., Souery, D., Montgomery, S., Albani, D., Forloni, G., Ferentinos, P., Rujescu, D., Mendlewicz, J., De Ronchi, D., Riva, M. A., Lewis, C. M., & Serretti, A. (2021). Drug repositioning for treatment-resistant depression: Hypotheses from a pharmacogenomic study. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 104, 110050. https://doi.org/10.1016/j.pnpbp.2020.110050
- Feng, H., Jiang, J., & Wei, G.-W. (2023). Machine-learning repurposing of DrugBank compounds for opioid use disorder. Computers in Biology and Medicine, 160, 106921. https://doi.org/10.1016/j.compbiomed.2023.106921
- Freshour, S. L., Kiwala, S., Cotto, K. C., Coffman, A. C., McMichael, J. F., Song, J. J., Griffith, M., Griffith, O. L., & Wagner, A. H. (2021). Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research, 49(D1), D1144–D1151. https://doi.org/10.1093/nar/gkaa1084
- Friedman, Y. (2023). DrugPatentWatch [Computer software]. thinkBiotech. www.DrugPatentWatch.com
- Gaulton, A., Hersey, A., Nowotka, M., Bento, A. P., Chambers, J., Mendez, D., Mutowo, P., Atkinson, F., Bellis, L. J., Cibrián-Uhalte, E., Davies, M., Dedman, N., Karlsson, A., Magariños, M. P., Overington, J. P., Papadatos, G., Smit, I., & Leach, A. R. (2017). The Chembl Database in 2017. Nucleic Acids Research, 45(D1), D945–D954. https://doi.org/10.1093/nar/gkw1074
- Hernández-Lemus, E., & Martínez-García, M. (2021). Pathway-Based Drug-Repurposing Schemes in Cancer: The Role of Translational Bioinformatics. Frontiers in Oncology, 10, 605680. https://doi.org/10.3389/fonc.2020.605680
- Hikmah, D. N., Syarifuddin, A., Santoso, S. B., Wijayatri, R., & Hidayat, I. W. (2024). The Role of Genetic Mutation on Schizophrenia: A Basic Review Prior to Pharmacogenomics. In Proceedings of the 4th Borobudur International Symposium on Humanities and Social Science 2022 (BIS-HSS 2022) (Vol. 778, pp. 835–847). Atlantis Press SARL. https://doi.org/10.2991/978-2-38476-118-0_96
- Irham, L. M., Adikusuma, W., Perwitasari, D. A., Dania, H., Maliza, R., Faridah, I. N., Santri, I. N., Phiri, Y. V. A., & Cheung, R. (2022). The use of genomic variants to drive drug repurposing for chronic hepatitis B. Biochemistry and Biophysics Reports, 31, 101307. https://doi.org/10.1016/j.bbrep.2022.101307
- Isik, Z., Baldow, C., Cannistraci, C. V., & Schroeder, M. (2015). Drug target prioritization by perturbed gene expression and network information. Scientific Reports, 5(1), 17417. https://doi.org/10.1038/srep17417
- Jarada, T. N., Rokne, J. G., & Alhajj, R. (2020). A Review of Computational Drug Repositioning: Strategies, Approaches, Opportunities, Challenges, and Directions. Journal of Cheminformatics, 12(1), 46. https://doi.org/10.1186/s13321-020-00450-7
- Jourdan, J.-P., Bureau, R., Rochais, C., & Dallemagne, P. (2020). Drug repositioning: A brief overview. Journal of Pharmacy and Pharmacology, 72(9), 1145–1151. https://doi.org/10.1111/jphp.13273
- Kim, S. (2021). Exploring Chemical Information in PubChem. Current Protocols, 1(8). https://doi.org/10.1002/cpz1.217
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., & Bolton, E. E. (2021). Pubchem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Research, 49(D1), D1388–D1395. https://doi.org/10.1093/nar/gkaa971
- Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B. A., Wang, J., Yu, B., Zhang, J., & Bryant, S. H. (2016). Pubchem Substance and Compound Databases. Nucleic Acids Research, 44(D1), D1202–D1213. https://doi.org/10.1093/nar/gkv951
- Krishnamurthy, N., Grimshaw, A. A., Axson, S. A., Choe, S. H., & Miller, J. E. (2022). Drug repurposing: A systematic review on root causes, barriers and facilitators. BMC Health Services Research, 22(1), 970. https://doi.org/10.1186/s12913-022-08272-z
- Kunz, M., Liang, C., Nilla, S., Cecil, A., & Dandekar, T. (2016). The drug-minded protein interaction database (DrumPID) for efficient target analysis and drug development. Database, 2016, baw041. https://doi.org/10.1093/database/baw041
- Lahti, J. L., Tang, G. W., Capriotti, E., Liu, T., & Altman, R. B. (2012). Bioinformatics and Variability in Drug Response: A Protein Structural Perspective. Journal of The Royal Society Interface, 9(72), 1409–1437. https://doi.org/10.1098/rsif.2011.0843
- Lauria, A., Mannino, S., Gentile, C., Mannino, G., Martorana, A., & Peri, D. (2020). DRUDIT: Web-Based Drugs Discovery Tools to Design Small Molecules as Modulators of Biological Targets. Bioinformatics, 36(5), 1562–1569. https://doi.org/10.1093/bioinformatics/btz783
- Lee, C. W., Kim, S. M., Sa, S., Hong, M., Nam, S.-M., & Han, H. W. (2023). Relationship between drug targets and drug-signature networks: A network-based genome-wide landscape. BMC Medical Genomics, 16(1), 17. https://doi.org/10.1186/s12920-023-01444-8
- Lesmana, M. H. S., Le, N. Q. K., Chiu, W.-C., Chung, K.-H., Wang, C.-Y., Irham, L. M., & Chung, M.-H. (2022). Genomic-Analysis-Oriented Drug Repurposing in the Search for Novel Antidepressants. Biomedicines, 10(8), 1947. https://doi.org/10.3390/biomedicines10081947
- Li, Y., Dong, Y., Qian, Y., Yu, L., Wen, W., Cui, X., & Wang, H. (2021). Identification of Important Genes and Drug Repurposing Based on Clinical-Centered Analysis Across Human Cancers. Acta Pharmacologica Sinica, 42(2), 282–289. https://doi.org/10.1038/s41401-020-0451-1
- Mendez, D., Gaulton, A., Bento, A. P., Chambers, J., De Veij, M., Félix, E., Magariños, M. P., Mosquera, J. F., Mutowo, P., Nowotka, M., Gordillo-Marañón, M., Hunter, F., Junco, L., Mugumbate, G., Rodriguez-Lopez, M., Atkinson, F., Bosc, N., Radoux, C. J., Segura-Cabrera, A., … Leach, A. R. (2019). ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Research, 47(D1), D930–D940. https://doi.org/10.1093/nar/gky1075
- Mensa-Wilmot, K. (2021). How Physiologic Targets Can Be Distinguished from Drug-Binding Proteins. Molecular Pharmacology, 100(1), 1–6. https://doi.org/10.1124/molpharm.120.000186
- Muresan, S., Sitzmann, M., & Southan, C. (2012). Mapping Between Databases of Compounds and Protein Targets. In R. S. Larson (Ed.), Bioinformatics and Drug Discovery (Vol. 910, pp. 145–164). Humana Press. https://doi.org/10.1007/978-1-61779-965-5_8
- Nam, Y., Kim, M., Chang, H.-S., & Shin, H. (2019). Drug Repurposing with Network Reinforcement. BMC Bioinformatics, 20(S13), 383. https://doi.org/10.1186/s12859-019-2858-6
- Napolitano, F., Carrella, D., Mandriani, B., Pisonero-Vaquero, S., Sirci, F., Medina, D. L., Brunetti-Pierri, N., & Di Bernardo, D. (2018). Gene2drug: A Computational Tool for Pathway-Based Rational Drug Repositioning. Bioinformatics, 34(9), 1498–1505. https://doi.org/10.1093/bioinformatics/btx800
- Ozery-Flato, M., Goldschmidt, Y., Shaham, O., Ravid, S., & Yanover, C. (2021). Framework for identifying drug repurposing candidates from observational healthcare data. JAMIA Open, 3(4), 536–544. https://doi.org/10.1093/jamiaopen/ooaa048
- Pabon, N. A., Xia, Y., Estabrooks, S. K., Ye, Z., Herbrand, A. K., Süß, E., Biondi, R. M., Assimon, V. A., Gestwicki, J. E., Brodsky, J. L., Camacho, C. J., & Bar-Joseph, Z. (2018). Predicting protein targets for drug-like compounds using transcriptomics. PLOS Computational Biology, 14(12), e1006651. https://doi.org/10.1371/journal.pcbi.1006651
- Parisi, D., Adasme, M. F., Sveshnikova, A., Bolz, S. N., Moreau, Y., & Schroeder, M. (2020). Drug Repositioning or Target Repositioning: A Structural Perspective of Drug-Target-Indication Relationship for Available Repurposed Drugs. Computational and Structural Biotechnology Journal, 18, 1043–1055. https://doi.org/10.1016/j.csbj.2020.04.004
- Park, K. (2021). The use of real-world data in drug repurposing. Translational and Clinical Pharmacology, 29(3), 117. https://doi.org/10.12793/tcp.2021.29.e18
- Peters, L., Rice, R., & Bodenreider, O. (2018). Rxnav-in-a-Box – a Locally-Installable Version of Rxnav and Related Apis. 2102.
- Phatak, S. S., & Zhang, S. (2013). A Novel Multi-Modal Drug Repurposing Approach for Identification of Potent ACK1 Inhibitors. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, 29–40.
- Pulley, J. M., Rhoads, J. P., Jerome, R. N., Challa, A. P., Erreger, K. B., Joly, M. M., Lavieri, R. R., Perry, K. E., Zaleski, N. M., Shirey-Rice, J. K., & Aronoff, D. M. (2020). Using What We Already Have: Uncovering New Drug Repurposing Strategies in Existing Omics Data. Annual Review of Pharmacology and Toxicology, 60(1), 333–352.
- https://doi.org/10.1146/annurev-pharmtox-010919-023537
- Quan, Y., Luo, Z.-H., Yang, Q.-Y., Li, J., Zhu, Q., Liu, Y.-M., Lv, B.-M., Cui, Z.-J., Qin, X., Xu, Y.-H., Zhu, L.-D., & Zhang, H.-Y. (2019). Systems Chemical Genetics-Based Drug Discovery: Prioritizing Agents Targeting Multiple/Reliable Disease-Associated Genes as Drug Candidates. Frontiers in Genetics, 10, 474. https://doi.org/10.3389/fgene.2019.00474
- Ramharack, P., & Soliman, M. E. S. (2018). Bioinformatics-Based Tools in Drug Discovery: The Cartography from Single Gene to Integrative Biological Networks. Drug Discovery Today, 23(9), 1658–1665. https://doi.org/10.1016/j.drudis.2018.05.041
- Romano, J. D., & Tatonetti, N. P. (2019). Informatics and Computational Methods in Natural Product Drug Discovery: A Review and Perspectives. Frontiers in Genetics, 10, 368. https://doi.org/10.3389/fgene.2019.00368
- Saberian, N., Peyvandipour, A., Donato, M., Ansari, S., & Draghici, S. (2019). A New Computational Drug Repurposing Method Using Established Disease–Drug Pair Knowledge. Bioinformatics, 35(19), 3672–3678. https://doi.org/10.1093/bioinformatics/btz156
- Sam, E., & Athri, P. (2019). Web-Based Drug Repurposing Tools: A Survey. Briefings in Bioinformatics, 20(1), 299–316. https://doi.org/10.1093/bib/bbx125
- Santoso, S. B., Chabibah, P. U., & Pribadi, P. (2021). Hepatotoxicity Risk Profile of Indonesian Due to Polymorphism of NAT2 and CYP2E1 in Isoniazid Metabolism. Urecol Journal. Part D: Applied Sciences, 1(1), 9–16. https://doi.org/10.53017/ujas.11
- Santoso, S. B., Pribadi, P., & Irham, L. M. (2023). Isoniazid-induced liver injury risk level in different variants of N-acetyltransferase 2 (NAT2) polymorphisms: A literature review. Pharmacia, 70(4), 973–981. https://doi.org/10.3897/pharmacia.70.e109869
- Setiyaningsih, H., Hidayat, I. W., Syarifuddin, A., Santoso, S. B., & Wijayatri, R. (2024). The Ruling of Novel Drugs to the Molecular Targets on Variant Pathological. In Proceedings of 5th Borobudur International Symposium on Humanities and Social Science (BISHSS 2023) (Vol. 856, pp. 4–13). Atlantis Press SARL. https://doi.org/10.2991/978-2-38476-273-6_2
- Sushko, I., Novotarskyi, S., Körner, R., Pandey, A. K., Rupp, M., Teetz, W., Brandmaier, S., Abdelaziz, A., Prokopenko, V. V., Tanchuk, V. Y., Todeschini, R., Varnek, A., Marcou, G., Ertl, P., Potemkin, V., Grishina, M., Gasteiger, J., Schwab, C., Baskin, I. I., Tetko, I. V. (2011). Online Chemical Modeling Environment (OCHEM): Web Platform for Data Storage, Model Development and Publishing of Chemical Information. Journal of Computer-Aided Molecular Design, 25(6), 533–554. https://doi.org/10.1007/s10822-011-9440-2
- Tse, T., Fain, K. M., & Zarin, D. A. (2018). How to Avoid Common Problems When Using Clinicaltrials.gov in Research: 10 Issues to Consider. BMJ, k1452. https://doi.org/10.1136/bmj.k1452
- Vogrinc, D., & Kunej, T. (2017). Drug Repositioning: Computational Approaches and Research Examples Classified According to the Evidence Level. Discoveries, 5(2), e75. https://doi.org/10.15190/d.2017.5
- Wang, K., Sun, J., Zhou, S., Wan, C., Qin, S., Li, C., He, L., & Yang, L. (2013). Prediction of Drug-Target Interactions for Drug Repositioning Only Based on Genomic Expression Similarity. PLoS Computational Biology, 9(11), e1003315. https://doi.org/10.1371/journal.pcbi.1003315
- Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., Wilson, M. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46(D1), D1074–D1082. https://doi.org/10.1093/nar/gkx1037
- Wishart, D. S., & Wu, A. (2016). Using DrugBank for In Silico Drug Exploration and Discovery. Current Protocols in Bioinformatics, 54(1). https://doi.org/10.1002/cpbi.1
- Wu, P., Feng, Q., Kerchberger, V. E., Nelson, S. D., Chen, Q., Li, B., Edwards, T. L., Cox, N. J., Phillips, E. J., Stein, C. M., Roden, D. M., Denny, J. C., & Wei, W.-Q. (2022). Integrating Gene Expression and Clinical Data to Identify Drug Repurposing Candidates for Hyperlipidemia and Hypertension. Nature Communications, 13(1), 46. https://doi.org/10.1038/s41467-021-27751-1
- Xia, X. (2017). Bioinformatics and Drug Discovery. Current Topics in Medicinal Chemistry, 17(15), 1709–1726. https://doi.org/10.2174/1568026617666161116143440
- You, J., McLeod, R. D., & Hu, P. (2019). Predicting drug-target interaction network using deep learning model. Computational Biology and Chemistry, 80, 90–101. https://doi.org/10.1016/j.compbiolchem.2019.03.016
- Zahrah, S. S., Wijayatri, R., Hidayat, I. W., Syarifuddin, A., & Santoso, S. B. (2024). From the Genetic Mutation to the Specific Pathologies. In Proceedings of the 4th Borobudur International Symposium on Humanities and Social Science 2022 (BIS-HSS 2022) (Vol. 778, pp. 439–447). Atlantis Press SARL. https://doi.org/10.2991/978-2-38476-118-0_49
- Zarin, D. A., Fain, K. M., Dobbins, H. D., Tse, T., & Williams, R. J. (2019). 10-Year Update on Study Results Submitted to ClinicalTrials.gov. New England Journal of Medicine, 381(20), 1966–1974. https://doi.org/10.1056/NEJMsr1907644
- Zhang, Y., Qazi, S., & Raza, K. (2021). Differential expression analysis in ovarian cancer: A functional genomics and systems biology approach. Saudi Journal of Biological Sciences, 28(7), 4069–4081. https://doi.org/10.1016/j.sjbs.2021.04.022
- Zheng, D. (2016). Pattern and application of tele-pharmacy service. Chinese Pharmaceutical Journal, 51(6), 513–518. https://doi.org/10.11669/cpj.2016.06.020
- Zhu, Y., Elemento, O., Pathak, J., & Wang, F. (2019). Drug knowledge bases and their applications in biomedical informatics research. Briefings in Bioinformatics, 20(4), 1308–1321. https://doi.org/10.1093/bib/bbx169
- Zong, N., Wen, A., Moon, S., Fu, S., Wang, L., Zhao, Y., Yu, Y., Huang, M., Wang, Y., Zheng, G., Mielke, M. M., Cerhan, J. R., & Liu, H. (2022). Computational drug repurposing based on electronic health records: A scoping review. Npj Digital Medicine, 5(1), 77. https://doi.org/10.1038/s41746-022-00617-6